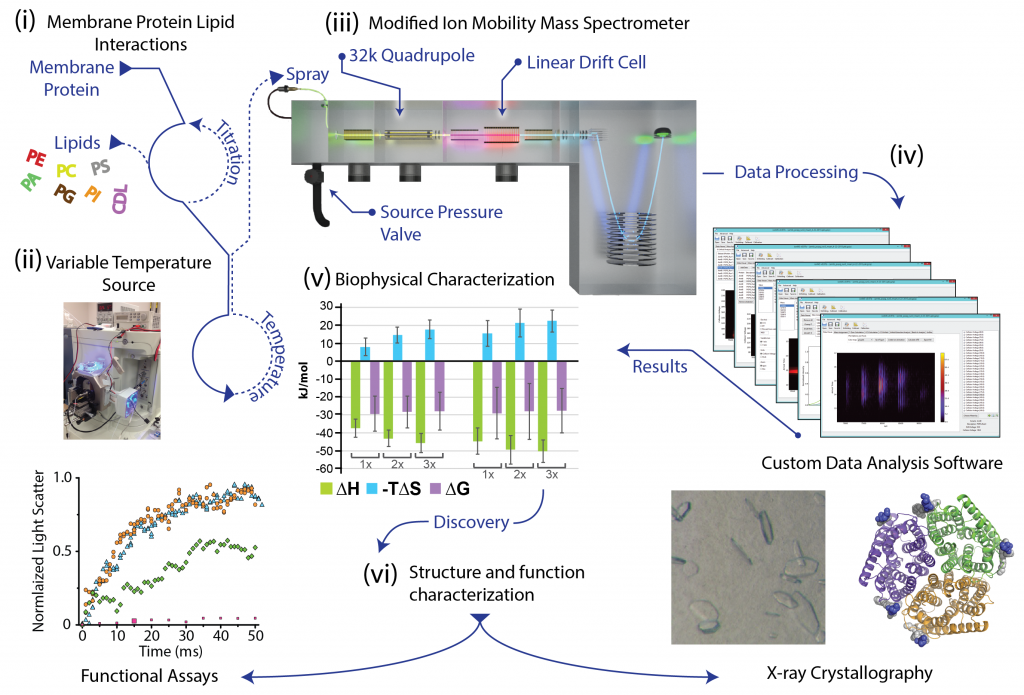
Membrane proteins are embedded in the biological membrane where they intimately interact with lipids. They represent one of the most important targets for pharmaceutical drug discovery, with a staggering 60% of drugs on the market targeting integral membrane proteins.[2, 3] The membrane environment is composed of a rich repertoire of chemically diverse lipids, with over 40,000 biologically relevant structures in the LIPID MAPS Structure Database to date.[4, 5] This striking number is over seven-fold more than the predicted human membrane proteome![8] Moreover, the lipid environment is dynamic, for example bacteria, in response to temperature, alter the degree of saturation and acyl length of their lipids to adjust membrane fluidity.[10] In the case of eukaryotic cells, lipids are distributed and maintained asymmetrically among the leaflets of organelles.[13] Lipids are also highly organized within membranes forming distinct domains, for example caveolae,[14] and micro domains, for example lipid rafts,[15, 16] that are enriched in specific lipids with unique functional properties. Despite the growing realization of the complexity of the biological membrane, we actually know little about how lipid molecules influence the structure and function of membrane proteins on the molecular level.
The crucial role of lipids in the folding, structure, and function of membrane proteins is emerging from exceedingly more reports.[17-23] However, a long-standing problem in this area of membrane protein biology has been the lack of technology to investigate membrane protein-lipid interactions. Of late, my laboratory has been developing new and innovative approaches to interrogate membrane protein-lipid interactions down to the resolution of individual lipid binding events using native ion mobility mass spectrometry (IM-MS), a technique where non-covalent interactions are preserved in the mass spectrometer.[17] Recently, we have developed cutting-edge approaches to obtain for the first time thermodynamic binding parameters for individual lipid binding events to membrane proteins and the ability to interrogate heterogeneous lipid binding events to membrane proteins at the resolution of individual lipid molecules [24]. In short, we are developing new and innovative native ion mobility mass spectrometry methods to address key questions in the field that remain otherwise intractable using other biophysical methods.
A long-term research goal of our group is to determine the molecular basis behind protein-lipid interactions and how these interactions can modulate the structure and function of membrane proteins, including their interactions with signaling molecules. What determines the selectivity of membrane proteins towards lipids, and the coupling between lipid binding events and function remains a key knowledge gap in the field; one that if addressed will significantly advance our understanding of how lipids participate in both normal and pathophysiological processes of membrane proteins. Therefore, there is a critical need to expand our fundamental knowledge in this emerging field by applying and developing innovative approaches to elucidate how lipids modulate the structure function of membrane proteins. To this end, we are studying a number of ion channels, receptors and other types of membrane proteins.